GenomeTrakr Fast Facts
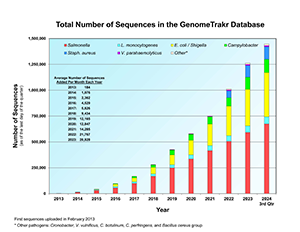
Total Number of Sequences in the GenomeTrakr Database (PDF: 96KB)
Basic Data Flow for Global WGS Public Access Databases (PDF: 61.7KB)
- GenomeTrakr is the first distributed network of labs to utilize whole genome sequencing for pathogen identification.
- Consists of 19 federal labs, 40 state health and university labs, 1 U.S. hospital lab, 2 other labs located in the U.S., 24 labs located outside of the U.S., and collaborations with independent academic researchers. More GenomeTrakr labs will be coming on-line.
- Data curation and bioinformatic analyses and support are provided by the National Center for Biotechnology Information (NCBI) at the National Institutes of Health.
- The GenomeTrakr network has sequenced more than 1.4 million isolates and closed more than 500 genomes. The network is regularly sequencing over 15,000 isolates each month.
- WGS can clearly define foodborne illness outbreaks.
- WGS networks are reliable, efficient, and can provide location specificity for outbreak investigations.
- Whole genome sequencing is inexpensive, easy to use, has identical sample prep for all pathogens, is the most accurate and high resolution subtyping technique, and a single test yields information about resistance, serotype, virulence factors, etc.
- The need for increased number of well characterized environmental (food, water, facility, etc.) sequences may outweigh the need for extensive clinical isolates.
- WGS programs can provide annual public health savings that far exceed the cost of funding such programs. In the U.S. the net benefits are estimated to be in the hundreds of millions of dollars annually and include reductions in the number of illnesses associated with the pathogens being sequenced.
- Basic Data Flow for Global WGS Public Access Databases (PDF: 4.29MB)