2021 FDA Science Forum
VICTRE 1.0+: Improved Computational Tools for Performing In Silico Imaging Clinical Trials
- Authors:
- Center:
-
Contributing OfficeCenter for Devices and Radiological Health
Abstract
Background
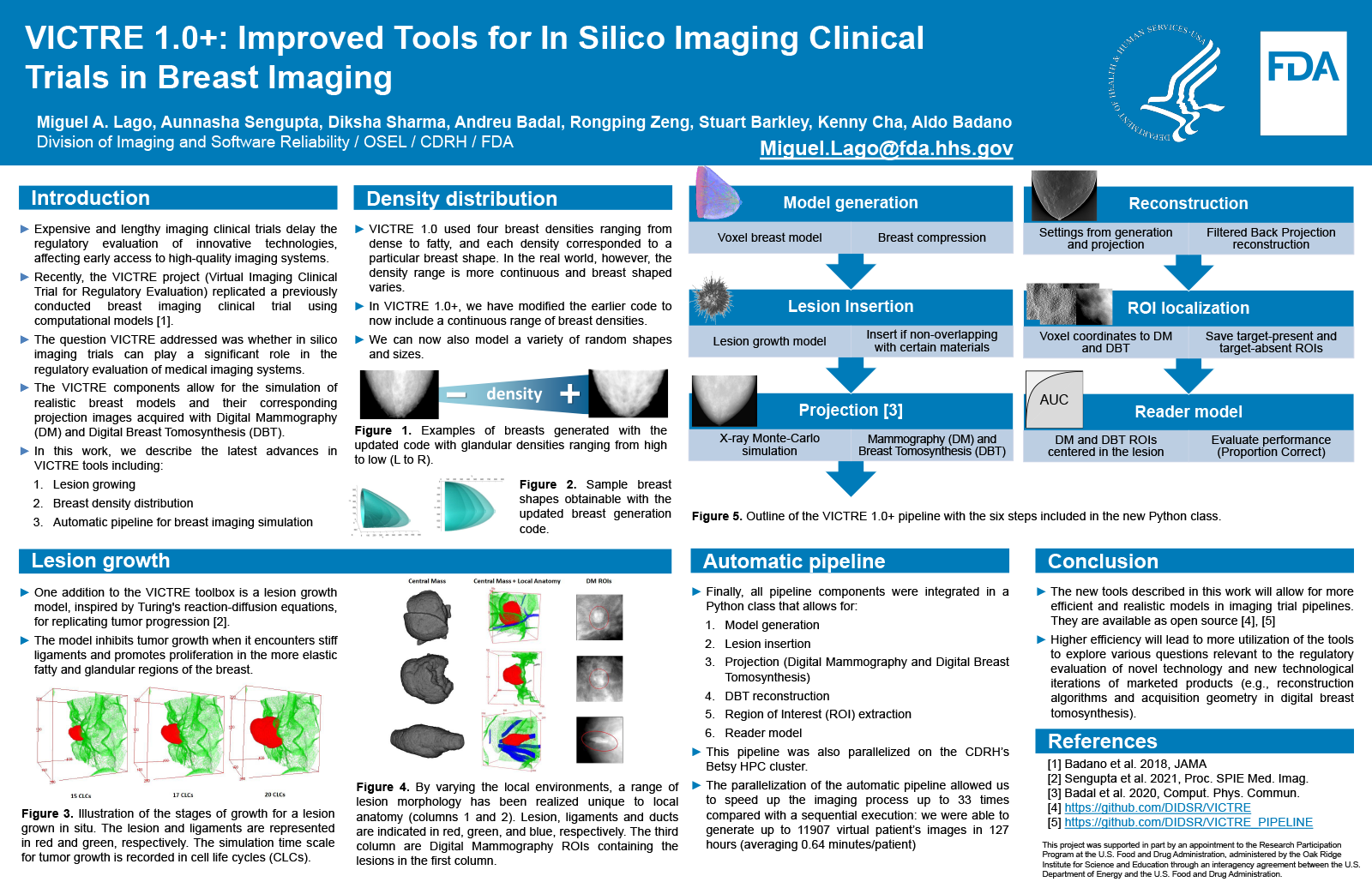
Expensive and lengthy imaging clinical trials delay the regulatory evaluation of innovative technologies, affecting early access to high-quality imaging systems. Recently, the VICTRE project (Virtual Imaging Clinical Trial for Regulatory Evaluation) replicated a previously conducted breast imaging clinical trial using computational models. The question VICTRE addressed was whether in silico imaging trials can play a significant role in the regulatory evaluation of medical imaging systems.
Purpose
In this work, we describe the latest advances in VICTRE tools including lesion growing, breast density, and an automatic version of the pipeline for breast imaging.
Methodology
One addition to the VICTRE toolbox is a lesion growth model, inspired by Turing's reaction-diffusion equations, for replicating tumor progression. The model inhibits tumor growth when it encounters stiff ligaments and promotes proliferation in the more elastic fatty and glandular regions of the breast. We have also developed code to obtain a distribution of radiographic density in a continuous space of breast shapes that will allow the accrual of virtual patient's with a range of characteristics. Finally, all pipeline components were integrated in a Python class that allows: 1) lesion insertion, 2) imaging simulation, 3) 3D reconstruction and 4) ROI extraction. This pipeline was also parallelized on the CDRH HPC computational cluster.
Results
By varying the environment where a lesions grows, a range of lesion morphology has been realized unique to local anatomy. The growth model was integrated with the pipeline to obtain x-ray breast images at several time points during the progression of disease. The parallelization of the automatic pipeline allowed us to speed up the imaging process up to 33 times compared with a sequential execution: we were able to generate up to 11907 patient’s images in 127 hours (averaging 0.64 minutes/patient).
Conclusion
The new tools described in this work will allow for more efficient and more realistic models used in imaging trial pipelines. Higher efficiency will lead to more utilization of the tools to explore various questions relevant to the regulatory evaluation of novel technology and new technological iterations of marketed products (e.g., reconstruction algorithms and acquisition geometry in digital breast tomosynthesis).