2023 FDA Omics Days - Poster Gallery
Poster Abstract Booklet
Poster Gallery
| Poster PDF | Title | Authors | Contact |
|---|---|---|---|
| Omics technologies to advance the FDA’s One Health Initiative | |||
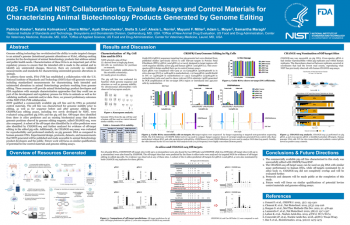
| FDA and NIST collaboration to evaluate assays and control materials for characterizing animal biotechnology products generated by genome editing | Patricia Kiesler, NIST; Natalia Kolmakova, NIST; Sierra Miller, NIST; Ayah Shevchenko, NIST; Stella S. Lee, FDA/CVM; Alexis L. Norris, FDA/CVM; Mayumi F. Miller, FDA/CVM; Adam L. Moyer, FDA/CVM; Samantha Maragh, NIST | patricia.kiesler@nist.gov | |
| RIPS: routine intuitive pathogen surveillance with whole genome sequence data | Tim Muruvanda, FDA/CFSAN; Arthur Pightling; FDA/CFSAN; James Pettengill, FDA/CFSAN; Hugh Rand, FDA/CFSAN | Tim.Muruvanda@fda.hhs.gov | |
| Genomics, transcriptomics, and metagenomics | |||
| Evaluating Renal Pathology in Post-COVID-19 Human Autopsy Tissues | Elysia Masters, FDA/NCTR; Mallikarjun Bidarimath, FDA/NCTR; Jennifer Hanks, TPA Inc.; Vaunita Parihar, Emory University; Amy Inselman, FDA/NCTR; Jessica Hawes, FDA/NCTR; Laura Schnackenberg, FDA/NCT; Kelly Mercer, FDA/NCTR | Elysia.Masters@fda.hhs.gov | |
| Comparison of target amplicon sequencing using the MiSeq and GridION next generation sequencing platforms for detection of foodborne pathogens | Isha Patel, FDA/CFSAN; Mark Mammel, FDA/CFSAN; Jayanthi Gangiredla, FDA/CFSAN | Isha.Patel@fda.hhs.gov | |
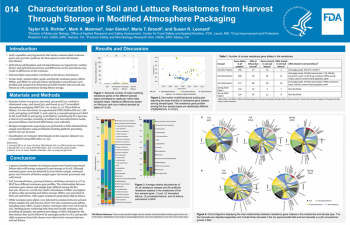
| Characterization of Soil and Lettuce Resistomes from Harvest Through Storage in Modified Atmosphere Packaging | Taylor K.S. Richter, FDA/CFSAN; Mark K. Mammel, FDA/CFSAN; Ivan Simko, USDA/ARS; Maria T Brandl, USDA/ARS; Susan R. Leonard, FDA/CFSAN | Taylor.Richter@fda.hhs.gov | |
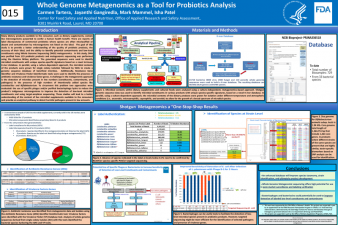
| Whole Genome Metagenomics as a Tool for Probiotics Analysis | Carmen Tartera, FDA/CFSAN; Jayanthi Gangiredla, FDA/CFSAN; Mark Mammel, FDA/CFSAN; Isha Patel, FDA/CFSAN | Carmen.Tartera@fda.hhs.gov | |
| Utility of miROmics for Identification of Circulating Pharmacodynamic Biomarkers of IFNβ-1a Biologics | Mai Mehanna, FDA/DARS; Lakshmi Manasa Chekka, FDA/DARS; Deepti Samarth, FDA/DARS; Erica Decker, DARS/FDA; Esraa Mohamed, FDA/DARS; Yan Guo, FDA/DARS; James L Weaver, DARS/FDA; Sarah Schrieber, OND/FDA; Jeffry Florian, FDA/DARS; Yow-Ming Wang, OCP/FDA; David Strauss, FDA/DARS; Paula Hyland, FDA/DARS | Mai.Mehanna@fda.hhs.gov | |
| PrecisionFDA Truth Challenge V2: Using Crowdsourcing to Benchmark Variant Calling Innovation | Ezekiel Maier, Booz Allen Hamilton; Elaine Johanson, FDA/ODT; Nathan Olson, NIST; Justin Wagner, NIST; Jennifer McDaniel, NIST; Justin Zook, NIST; Samuel Westreich, DNAnexus; Omar Serang, DNAnexus; Anish Prasanna, Booz Allen Hamilton | Adrienne.Phifer@fda.hhs.gov | |
| Assessment of plasmids for relating the 2020 Salmonella enterica serovar Newport onion outbreak to farms implicated by the outbreak investigation | Seth Commichaux, FDA/CFSAN; Hugh Rand, FDA/CFSAN; Kiran Javkar, University of Maryland/Department of Computer Science; Erin K. Molloy, University of Maryland/Department of Computer Science; James B. Pettengill, FDA/CFSAN; Arthur Pightling, FDA/CFSAN; Maria Hoffmann, FDA/CFSAN; Mihai Pop, University of Maryland/Department of Computer Science; Victor Jayeola, FDA/CFSAN; Steven Foley, FDA/NCTR; Yan Luo, FDA/CFSAN | Seth.Commichaux@fda.hhs.gov | |
| Global transcriptomic analyses of Salmonella enterica serovar Agona reveals the mechanism of survival on low moisture food | Sultana Solaiman, FDA/CFSAN; Ellie Meeks, UMD/JIFSAN; Ian Hines, FDA/CFSAN; Jie Zheng, FDA/CFSAN; Maria Hoffmann, FDA/CFSAN | Sultana.Solaiman@fda.hhs.gov | |
|
|
Altered transcriptome and chromatin dynamics following short- and long-term Zika infections | Aaron Scholl, FDA/CBER; Bingjie Li, FDA/CBER; Sandip De, FDA/CBER | Aaron.Scholl@fda.hhs.gov |
| Metagenomics and Targeted Capture Next Generation Sequencing to Detect and Identify Insects in Foods | Monica Pava-Ripoll, FDA/CFSAN/OFS; Mark K. Mammel, FDA/CFSAN/OARSA; Elizabeth Reed, FDA/CFSAN/ORS; Martine Ferguson, FDA/CFSAN/OAO; and Padmini Ramachandran, FDA/CFSAN/ORS | Monica.Pava-Ripoll@fda.hhs.gov | |
| Assessing viral variant detection algorithms to improve characterization of gene therapy products and biologics | Hiral Desai, MilliporeSigma; Yanfei Zhou, MilliporeSigma; Bradley Hassan, MilliporeSigma; Alexandra Bridgeland, MilliporeSigma; William Dolan Milliporesigma; Amber Overgard, MilliporeSigma | hiral.desai@milliporesigma.com | |
| Temporal changes in Shiga-Toxin producing Escherichia coli (STEC) O121 transcriptome during storage in bleached flour | Ian_Hines, FDA/CFSAN; Emily_Nguyen, UMD/JIFSAN; Ellie_Meeks, UMD/JIFSAN; Sultana_Solaiman, FDA/CFSAN; Elizabeth_Reed, FDA/CFSAN; Maria_Hoffmann, FDA/CFSAN; Jie_Zheng, FDA/CFSAN | Ian.Hines@fda.hhs.gov | |
|
Not Available
|
A Machine Learning Approach for Identifying Variables Associated with Risk of Developing Neutralizing Antidrug Antibodies to Factor VIII | Atul Rawal, CBER/OTP/HB; Christopher Kidchob, CBER/OTP/HB; Jiayi Ou, CBER/OTP/HB; Zuben E. Sauna, CBER/OTP/HB; Osman N. Yogurtcu, CBER/OBP ; Hong Yang, CBER/OBP | Atul.Rawal@fda.hhs.gov |
| Not Available | Single cell sequencing of brain sequestered CD8+ T cells during experimental cerebral malaria | Miranda S. Oakley/DETTD, CBER, FDA; Micah Phillip/ Emory University; Victoria Majam/DETTD, CBER, FDA; Hong Zheng/DETTD, CBER, FDA; Mark A. KuKuruga/DBPAP, CBER, FDA; Gregory K. Tharp/ Emory University; Sanjai Kumar/DETTD, CBER, FDA | Miranda.Oakley@fda.hhs.gov |
| The Transcriptome Landscape of 3D-cultured Placental Trophoblasts Reveals Activation of TLR2 and TLR3/7 in Response to Trypanosoma cruzi | Erica Silberstein, FDA/CBER/OBRR; Charles C. Chung, FDA/HIVE, Alain Debrabant, FDA/CBER/OBRR | Erica.Silberstein@fda.hhs.gov | |
| Dual scRNA-Seq Analysis Reveals Rare and Uncommon Parasitized Cell Populations in Chronic L. Donovani Infection | Thalia Pacheco-Fernandez FDA/CBER/OBRR/DETTD/LEP; Konstantinos Karagiannis PCV Bioinformatics Engineering, Moderna; Sreenivas Gannavaram FDA/CBER/OBRR/DETTD/LEP; Chaitenya Verma Wexner Medical Center, The Ohio State University; Parna Bhattacharya FDA/OHTVII/CDRH; Hira L. Nakhasi FDA/CBER/OBRR/DETTD/LEP; Abhay R. Satoskar Wexner Medical Center, The Ohio State University | Thalia.PachecoFernandez@fda.hhs.gov | |
| CSP2: A Nextflow Pipeline for the Fast and Accurate Genetic Distance Estimation from Assembled Pathogen Genomes | Robert Literman, FDA/CFSAN/OAO; James Pettengill, FDA/CFSAN/OAO; Hugh Rand, FDA/CFSAN/OAO | Robert.Literman@fda.hhs.gov | |
| Not Available | Analysis of Various Cryopreservation Conditions for the Storage of Induced Pluripotent Stem Cells and other Cell-based Therapies | Shweta Kotian, U.S. Food and Drug Administration (FDA); Farhad Farjood, Neural Stem Cell Institute (NSCI); Nathan Boles, Neural Stem Cell Institute (NSCI); John T. Thomas, U.S. Food and Drug Administration (FDA); Brigitte L. Arduini, Neural Stem Cell Institute (NSCI); Jeffrey Stern, Neural Stem Cell Institute (NSCI); Sally Temple, Neural Stem Cell Institute (NSCI); Malcolm Moos Jr., U.S. Food and Drug Administration (FDA) | Shweta.Kotian@fda.hhs.gov |
| Proteomics and metabolomics | |||
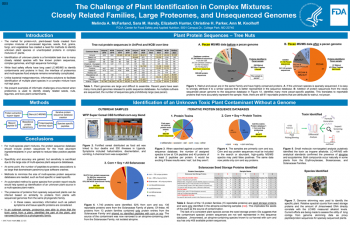
| The Challenge of Plant Identification in Complex Mixtures: Closely Related Families, Large Proteomes, and Unsequenced Genomes | Melinda A. McFarland, FDA/CFSAN/ORS; Sara M. Handy, FDA/CFSAN/ORS; Elizabeth Hunter, FDA/CFSAN/ORS; Christine H. Parker, FDA/CFSAN/ORS; Ann M. Knolhoff, FDA/CFSAN/ORS | Melinda.McFarland@fda.hhs.gov | |
| Proteomic Profiling Reveals Antibiotic Resistance Mechanisms in Staphylococcus epidermidis Biofilms under Tigecycline Pressure | Kidon Sung, FDA/NCTR; Miseon Park, FDA/NCTR; Jungwhan Chon, Kyungin Women's University, Korea; Ohgew Kweon, FDA/NCTR; Saeed Khan, FDA/NCTR; Andrew Shen, FDA/NCTR; Angel Paredes, FDA/NCTR | Kidon.Sung@fda.hhs.gov | |
| Optimizing dia-PASEF isolation window schemes for proteomics measurements on a timsTOF ultra instrument | Ryan Marsico, Bruker Daltonics; Markus Lubeck, Bruker Daltonics; ,Stephanie Kaspar-Schoenefeld, Bruker Daltonics; Christoph Krisp, Bruker Daltonics; Andreas Schmidt, Bruker Daltonics; Florian Busch, Bruker Daltonics; Eduardo Carrascosa, Bruker Daltonics; Oliver Raether; Gary Kruppa, Bruker Daltonics | Ryan.Marsico@bruker.com | |
|
Not Available
|
Deeper plasma proteome coverage enables identification of novel biomarkers and classification of diseases | Shourjo Ghose, Andreas Schmidt, Zehan Hu, Claudia Martelli, Katharina Limm, Xaver Wurzenberger, Katrin Hartinger, Sebastian Mueller, Nils A. Kulak | shourjo.ghose@bruker.com |
| Identifying metabolite markers for detection of flaviviruses in human induced Pluripotent Stem Cells (hiPSCs) | Tahira Fatima OTP/DCT2/TVBB; Sandip De OTP/DCT2/TVBB | Tahira.Fatima@fda.hhs.gov | |
|
|
A practical lock-mass calibrant introduction method for improved mass accuracy and reduced false positive identifications in non-targeted analyses | Christine Fisher (O'Donnell), FDA/CFSAN; Shannon Murphy, FDA/CFSAN; Ann Knolhoff, FDA/CFSAN | Christine.ODonnell@fda.hhs.gov |
| When is Enough Actually Enough- How Does the Number of Replicates Influence the Quality of Non-Targeted Analysis Results? | Karen Butler, FDA/CFSAN; Erica Bakota, FDA/ORA; Christine Fisher, FDA/CFSAN; Brian Ng, FDA/CFSAN; Ann Knolhoff, FDA/CFSAN | Karen.Butler@fda.hhs.gov | |
| Metabolomics Evaluation of the Impact of Violet-Blue Light (405 nm) on Platelet Concentrate | Chintamani D. Atreya, CBER/FDA; Jinchun Sun, FDA/NCTR; Neetu Dahiya, FDACBER, Tom Schmitt, FDA/NCTR; Caitlin Stewart, U of Strath; John Anderson; U of Strath; Scott MacGregor, U of Strath; Michelle Maclean, U of Strath; Richard. D. Beger, FDA/NCTR | Chintamani.Atreya@fda.hhs.gov | |
| Application of Metabolomic Analysis Towards the Discovery of Biomarkers of Immunogenecity and Efficacy of Parasitic Vaccines | Nazli Azodi, FDA/CBER/OBRR/DETTD/LEP; Hannah Markle, FDA/CBER/OBRR/DETTD/LEP; Timur Oljuskin, USDA; Parna Bhattacharya, FDA/OHTVII/CDRH; Greta Volpedo, The Ohio State University; Abhay Satoskar, The Ohio State University; Sreenivas Gannavaram, FDA/CBER/OBRR/DETTD/LEP; Hira L. Nakhasi, FDA/CBER/OBRR/DETTD/LEP | Sreenivas.Gannavaram@fda.hhs.gov | |
| Evaluation of plasma proteome and miRNA changes related to COVID-19 patient severity response | Beger, Richard, FDA/NCTR; Yu, Li-Rong FDA/NCTR; Han Tao, FDA/NCTR; Vijay, Vikrant, FDA/NCTR; Pence, Lisa, FDA/NCRT; Sun, Jinchun, FDA/NCTR; Bidarimath, Mallikarjun, FDA/NCTR; Schmitt, Thomas, FDA/NCTR; Mercer, Kelly, FDA/NCTR; Masters, Elysia, FDA/NCTR; Hawes, Jessica, FDA/NCTR; Smallwood, Heather, UTHSC; Arthur, John UAMS; Bhattacharya, Sudeepa, ASU; Burkhart, Keith, FDA/CDER | Richard.Beger@fda.hhs.gov | |
|
Not Available
|
Applying MHC-II Associated Peptide Proteomics (MAPPs) assay to study the Immunogenicity of Staphylococcus aureus Cas9 | Louis Hopkins, FDA/CBER; Joseph R. McGill, FDA/CBER; Swati Mukherjee, Editas; Kate Zhang, Editas; and Zuben E. Sauna,FDA/CBER; | Vijaya.Simhadri@fda.hhs.gov |
| Lipidomics Evaluation of the Impact of Fentanyl Treatments on Neural Stem Cells | Richard Beger, FDA/NCTR; Jinchun Sun, FDA/NCTR; Rohini Donakonda, FDA/NCTR; Shuliang Liu, FDA/NCTR; Fang Liu, FDA/NCTR; Cheng Wang, FDA/NCTR | Richard.Beger@fda.hhs.gov | |
| MSlineaR: a new tool to assess linearity, improving statistical robustness and quality assurance in untargeted metabolomics | Janine Wiebach, Metabolomics Platform, Berlin Institute of Health at Charité – Universitätsmedizin Berlin; Jennifer Kirwan, Metabolomics Platform, Berlin Institute of Health at Charité – Universitätsmedizin Berlin; Álvaro Fernández Ochoa, Department of Analytical Chemistry, Faculty of Sciences, University of Granada | Janine.wiebach@bih-charite.de | |
| Identification of Circulating Pharmacodynamic Biomarkers of IL-5 Inhibitors using proteomics approach | Samarth Deepti, FDA/CDER; Chekka Lakshmi Manasa, FDA/CDER; Mohammed Esraa, FDA/CDER; Decker Erica; Guo Yan; Wheeler Will, IMS Inc.; Howard Kristina, FDA/CDER; Schrieber Sarah J; Florian Jeffey FDA/CDER; Wang Yow-Ming, FDA/CDER; Strauss David G, FDA/CDER, Hyland Paula L, FDA/CDER | Deepti.Samarth@fda.hhs.gov | |
| Lysophosphatidylcholine mediates neutrophil activity through early metabolic modulation following immunization with a live-attenuated Leishmania vaccine | Hannah L Markle, FDA/CBER/OBRR/DETTD/LEP; Thalia Pacheco-Fernandez, FDA/CBER/OBRR/DETTD/LEP, Parna Bhattacharya, FDA/OHT7/CDRH; Jinchun Sun, FDA/NCTR; Nazli Azodi, FDA/CBER/OBRR/DETTD/LEP; Grace Kitthanawong, FDA/CBER/OBRR/DETTD/LEP; Caroline Hobson, FDA/CBER/OBRR/DETTD/LEP; Richard Beger, FDA/NCTR; Sreenivas Gannavaram, FDA/CBER/OBRR/DETTD/LEP, Hira Nakhasi, FDA/CBER/OBRR/DETTD/LEP | Hannah.Markle@fda.hhs.gov | |
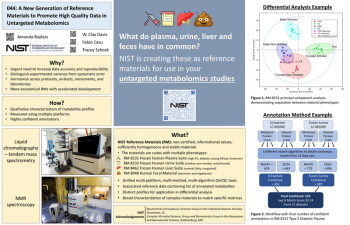
| A New Generation of Reference Materials to Promote High Quality Data in Untargeted Metabolomics | Amanda Bayless, NIST/CSD; Clay Davis, NIST/CSD; Fabio Casu, NIST/CSD; Tracey Schock, NIST/CSD | Amanda.bayless@nist.gov | |
| High resolution MALDI imaging mass spectrometry to assess spatial lipidomics of mouse fetal neural tube defects after maternal opioid exposure | Dustyn Barnette, FDA/NCTR, E. Ellen Jones, FDA/NCTR, Pravin Kaldhone, FDA/CBER, Grace Lee, FDA/CDER, Kelly Davis, FDA/NCTR, Sumit Sarkar, FDA/NCTR, Pritpal Malhi, FDA/CDER, J. Edward Fisher, FDA/CDER, Joseph P. Hanig, FDA/CDER, Richard D. Beger, FDA/NCTR, Richard D. Mellon, FDA/CDER, Amy L. Inselman, FDA/NCTR | Dustyn.Barnette@fda.hhs.gov | |
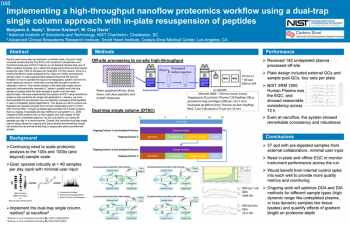
| Implementing a high-throughput nanoflow proteomics workflow using a dual-trap single column approach with in-plate resuspension of peptides | Benjamin Neely, NIST; W. Clay Davis, NIST | benjamin.neely@nist.gov | |
| Data integration and data management | |||
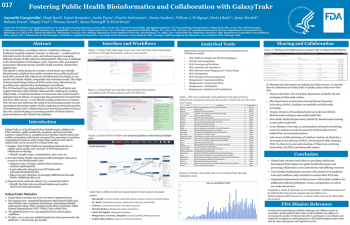
| Fostering Public Health Bioinformatics and Collaboration with GalaxyTrakr | Jayanthi Gangiredla, FDA/CFSAN; Hugh Rand, FDA/CFSAN; Daniel Benisatto, DRT Strategies; Justin Payne, FDA/CFSAN; Charles Strittmatter, FDA/CFSAN; Jimmy Sanders, SDS Solutions, Inc.; William J. Wolfgang, New York State Department of Health/Wadsworth Center; Kevin Libuit, Division of Consolidated Laboratory Services/Department of General Services; James Herrick, James Madison University/Biology Department ; Melanie Prarat, Ohio Department of Agriculture/Animal Disease Diagnostic Laboratory; Magaly Toro, Universidad de Chile/Laboratorio de Microbiología y Probióticos; Thomas Farrell, FDA/CFSAN; James Pettengill, FDA/CFSAN; Errol Strain, FDA/CFSAN; | Jayanthi.Gangiredla@fda.hhs.gov | |
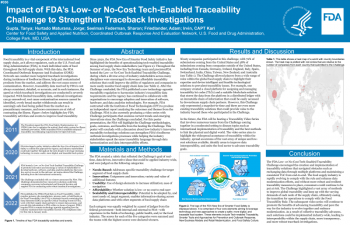
| Impact of FDA’s Low- or No-Cost Tech-Enabled Traceability Challenge on Strengthening Traceback Investigations | Tanya Gupta, FDA/CFSAN/OCORE; Jorge Hurtado Maturana, FDA/CFSAN/OCORE; Sharon Seelman, FDA/CFSAN/OCORE; Adam Friedlander, FDA/CFSAN/OCORE; CAPT Kari Irvin, FDA/CFSAN/OCORE | Tanya.Gupta@fda.hhs.gov | |