NARMS Interim Data Updates
What are NARMS Interim Data Updates?
NARMS interim data updates provide a summary of recent unusual or concerning antimicrobial resistance findings in enteric bacteria found through surveillance of retail meats. These updates are based upon using whole-genome sequence (WGS) analysis to screen for bacterial genes associated with resistance to one or more medically important antibiotics. Findings that would lead to a data update include but are not limited to
- Resistance to antimicrobials typically used to treat infections in humans (e.g., ceftriaxone and ciprofloxacin for treatment of severe Salmonella infections),
- Resistance to antimicrobials used as last-resort treatments (e.g., colistin), and
- Emergence of extensively drug-resistant clones.
NARMS interim data updates do not serve as notice of product recalls, alerts, or outbreaks of foodborne illness. Information about these events can be found at
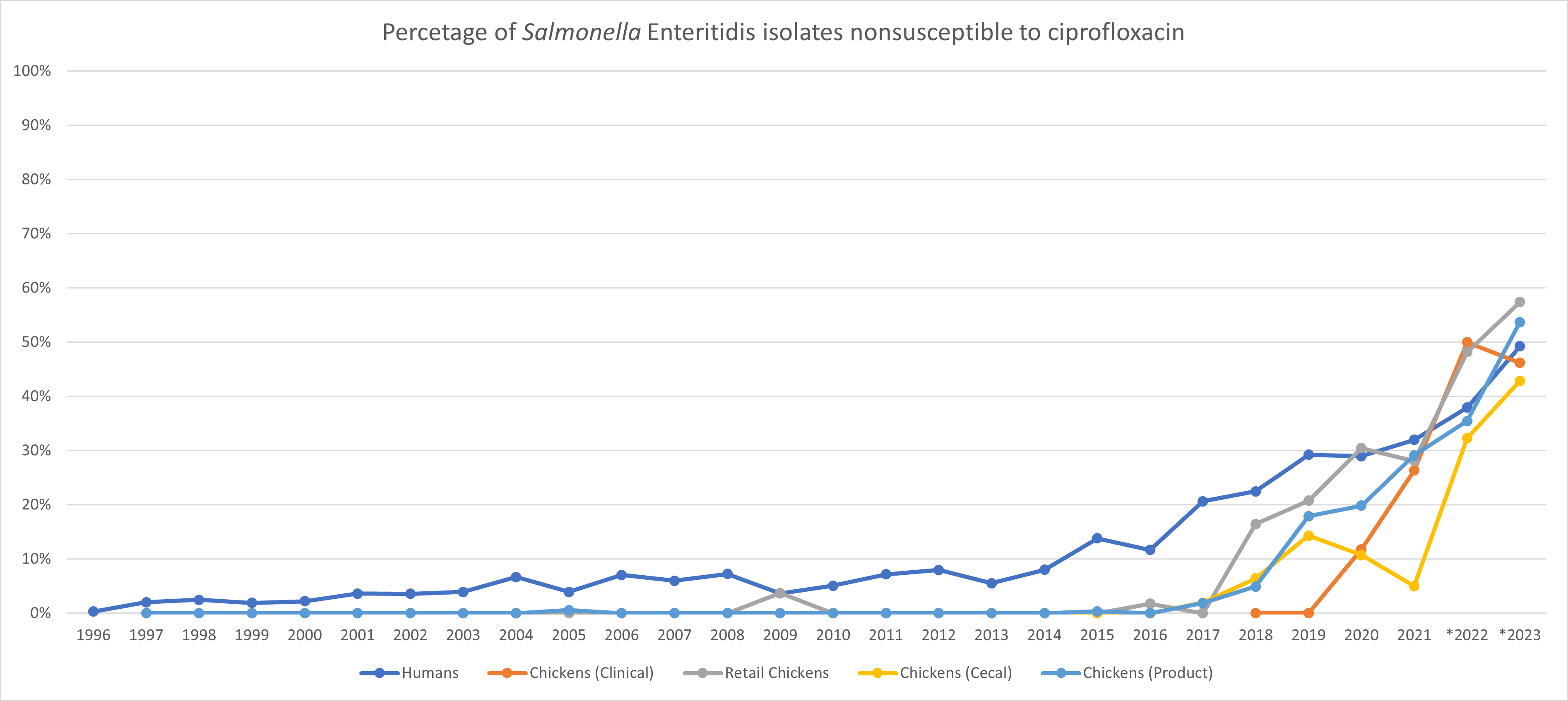
NARMS Interim Data Update: Increasing trend of Salmonella Enteritidis that have decreased susceptibility to ciprofloxacin
This interim update expands on observations highlighted in the 2021 National Antimicrobial Resistance Monitoring System (NARMS) Integrated Report Summary. In that summary, NARMS observed a rise in Salmonella serovar Enteritidis strains from ill persons, retail chicken, and chickens at slaughter that exhibit decreased susceptibility to ciprofloxacin (DSC, MIC ≥ 0.12 µg/mL). Although NARMS is not reporting an increase in Salmonella Enteritidis overall, an increase in DSC among Enteritidis is concerning because it could adversely affect response to treatment with ciprofloxacin, a fluoroquinolone class of antimicrobials commonly used to treat severe salmonellosis. CLSI defines Salmonella with ciprofloxacin MICs ≥0.12 µg/mL as being ‘Intermediate’ or ‘Resistant’; infections with these strains may be associated with clinical failure or delayed response in fluoroquinolone-treated patients. The Salmonella Enteritidis strains exhibiting the DSC phenotype commonly carry a mutation in the gyrase A (gyrA) gene that results in an amino acid change (D87Y) in the GyrA protein (the target of fluoroquinolones).
Source of animal data: NARMS Now Integrated Data
Source of human data: NARMS Now Human Data; *2022 and 2023 human data are preliminary
Of the DSC Salmonella Enteritidis isolates sequenced during 2017–2023, the D87Y mutation was seen in 99.4% (812/817) of isolates from chickens at slaughter and chicken meat, and in 74.4% (486/653) of human isolates tested for NARMS routine surveillance. The mutation was also present in all 22 DSC Salmonella Enteritidis isolates from sick chickens sampled by VetLIRN laboratories during 2018–2023. Most DSC Salmonella Enteritidis isolates with a D87Y mutation in GyrA harbor no additional resistance genes. SNP clustering analysis indicated that commercial chicken products are a likely source of these strains. These strains have also been linked to outbreaks associated with eggs in 2024 (93 cases) and backyard poultry (93 cases during the 2022 season and 41 cases during the 2024 season). Additionally, 7 investigations (2019–2024) involving multiple states have been linked to these strains and were attributed to international travel, mainly to the Caribbean region. These alternate sources and routes of exposure may contribute to additional cases of human illness caused by DSC Salmonella Enteritidis.
As fluoroquinolones have been prohibited for use in all U.S. poultry since 2005, the NARMS program is working with industry partners to further understand the origin and potential drivers of DSC Salmonella Enteritidis and also to explore mitigation strategies.
Consumer Safety
The purpose of this notification is to inform stakeholders of this emergent finding. The public is advised to continue practicing safe handling of raw meats, as recommended by the USDA, and how to stay healthy around backyard poultry, as recommended by the CDC.
NARMS Testing Interim Data Update: Salmonella I 4,[5],12:i:- with Azithromycin Resistance
New Finding
NARMS whole-genome sequencing (WGS) analysis indicates an increase in multidrug-resistant (MDR) strains of Salmonella I 4,[5],12:i:- (i.e., monophasic Salmonella serovar Typhimurium) in the United States. These strains have predicted azithromycin resistance (AZM-R) due to presence of the mph(A) gene. These strains of AZM-R I 4,[5],12:i:- have been isolated from 112 ill people from 34 states between 2017 and 2024, with the majority identified in 2023. Of the 15 ill people with available information, 9 were hospitalized. No deaths have been reported. The median age of ill people is 26 years (interquartile range 8–53.5 years) and 60% are male. These strains of AZM-R I 4,[5],12:i:- have also been isolated from 6 ill animals (1 calf, 1 horse, and 4 pigs) as a part of the Vet-LIRN antimicrobial resistance monitoring program, 3 animals at slaughter (swine cecal samples), and 6 food product (pork) samples from 2019 to 2024.
Number of MDR Salmonella I 4,[5],12:i:- isolates with azithromycin resistance gene mph(A) tested by NARMS, by isolation year, January 1, 2017–May 29, 2024
All 127 Salmonella I 4,[5],12:i:- isolates are MDR (predicted resistance to at least 3 classes of antimicrobials); 26% are extensively drug-resistant (XDR), here defined as predicted resistance to all commonly recommended empiric and alternative antimicrobials — ampicillin, azithromycin, ceftriaxone, ciprofloxacin, and trimethoprim-sulfamethoxazole.
Percentage of Salmonella I 4,[5],12:i:- isolates with azithromycin resistance gene mph(A) that were predicted resistant, by antimicrobial (n = 127), as of May 29, 2024 — NARMS
Note: Resistance is predicted based on the presence of at least one resistance determinant for each antimicrobial, identified through whole genome sequencing; includes isolates from human, food, and animal sources. Individual results can be found online: Isolates Browser - Pathogen Detection - NCBI (nih.gov)
* Extensively drug-resistant (XDR): predicted resistance to all commonly recommended empiric and alternative antibiotics for treatment of salmonellosis — ampicillin, azithromycin, ceftriaxone, ciprofloxacin (including predicted decreased susceptibility), and trimethoprim-sulfamethoxazole
**Ciprofloxacin includes isolates with at least one quinolone resistance gene or mutation predicting nonsusceptibility (a single mechanism is typically associated with an antimicrobial susceptibility testing interpretation of "intermediate" for ciprofloxacin in Salmonella).
Although these strains have not caused recognized outbreaks of related illness or been linked to specific sources in the United States, an mph(A)-positive strain of XDR Salmonella I 4,[5],12:i:- that is genetically related to a subset of these isolates has been reported in Canada by the Public Health Agency of Canada. The XDR I 4,[5],12:i:- strain reported in Canada was linked to raw pet food and contact with cattle (https://www.canada.ca/en/public-health/services/public-health-notices/2023/outbreak-salmonella-infections-under-investigation.html).
Caution Statement
An increase in AZM-R Salmonella I 4,[5],12:i:- with predicted resistance to critically important and other antimicrobials in humans and its occurrence in food-producing animals and foods is a public health concern.
Next Steps
NARMS agencies are performing additional work to better understand the origin and spread of these strains and the genes resulting in their MDR/XDR profiles.
Consumer Safety
This notification is to inform stakeholders of this emergent finding. The FDA, CDC, and USDA will continue to monitor the situation. The public is advised to continue practicing safe handling of raw meats, as recommended by the USDA, and proper hygiene around animals, as recommended by the CDC.
NARMS Retail Meat Testing Notification: Multidrug-Resistant (MDR) Salmonella Senftenberg
New Finding
NARMS sampling of retail ground turkey has identified a Salmonella serotype Senftenberg isolate (19GA12GT11-S) with a plasmid carrying a cephalosporin resistance gene. This plasmid is similar to the mega-plasmid (pESI) found in a multidrug-resistant Salmonella Infantis strain that has become increasingly prevalent in U.S. poultry and cases of human illnesses. This cephalosporin-resistant Senftenberg strain was first detected by NARMS in 2019 (1 sample) and again in 2021 (3 samples) and 2022 (3 samples), but the plasmid and its similarity to the pESI in multidrug-resistant Salmonella Infantis was only recently confirmed through further genomic analysis. CDC has identified two cases of human illness caused by this strain since June 2022. One patient reported consuming ground turkey in the seven days before illness began, and the other had no known turkey exposure.
This Salmonella serotype Senftenberg strain is resistant to 8 out of 14 antibiotics on the NARMS Salmonella panel (see Table). Ceftriaxone (a cephalosporin) is considered a critically important drug for treating severe Salmonella infections, especially for children. The development and spread of resistance to critically important antimicrobials is a serious public health concern (CDC, 2022). To increase public health awareness, the FDA is sharing this information via this notification, and the USDA and FDA have also proactively updated representatives from the turkey industry about this finding.
MDR profile of the NARMS isolate 19GA12GT11-S
| Class | Antimicrobial Agent | Resistant | Genes |
|---|---|---|---|
| Aminoglycosides | Gentamicin | X | aac(3)-IV |
| Aminoglycosides | Streptomycin | X | aadA1, aph(3')-Ia, aph(3'')-Ib |
| Beta-lactam combination agents | Amoxicillin-Clavulanic Acid | ||
| Cephems | Cefoxitin | ||
| Cephems | Ceftriaxone | X | blaCTX-M-65 |
| Folate pathway antagonists | Sulfisoxazole | X | sul1,sul2 |
| Folate pathway antagonists | Trimethoprim-Sulfamethoxazole | X | dfrA14; sul1,sul2 |
| Macrolides | Azithromycin | ||
| Penems | Meropenem | ||
| Penicillins | Ampicillin | X | blaCTX-M-65 |
| Phenicols | Chloramphenicol | X | floR |
| Quinolones | Ciprofloxacin | ||
| Quinolones | Nalidixic Acid | ||
| Tetracyclines | Tetracycline | X | tet(A) |
Fast Facts
Isolate(s): 19GA12GT11-S, 21NC10GT04-S, 21NY01GT05-S1, 21NM11GT05-S1, CFSAN119630, PNUSAS276269, 22MO06GT03-S1
Organism: Salmonella Senftenberg
Source: Retail ground turkey
Location(s): GA, MD, MO, NC, NM, NY, PA
Date collected: 2019, 2021, and 2022
Other strains from different lineages of Salmonella Senftenberg have been associated with outbreaks of human illness linked to pistachios (FDA Investigation) and to peanut butter (FDA Investigation). Salmonella Senftenberg has been routinely detected in NARMS retail testing of ground turkey (55 total isolates from 2016 to 2021 testing period). Additional information on Salmonella Senftenberg from turkey that are genetically similar to 19GA12GT11-S can be found on NCBI’s Pathogen Detection Portal.
Consumer Safety
This notification is to inform stakeholders of this novel finding. Although this rare finding by itself does not signify elevated risk, the public is advised to continue practicing safe handling of raw meats, as recommended by the USDA, and proper hygiene around animals as recommended by the CDC.
NARMS Retail Meat Testing Interim Data Update: Multidrug-Resistant (MDR) Salmonella I 4,[5],12:i:-
New Finding
In November of 2019, NARMS collected a retail pork sample that yielded a Salmonella serotype I 4,[5],12:i:- isolate (19MN11PC02) that is resistant to 10 out of 14 antibiotics on the NARMS Salmonella panel (see Table). Further genetic characterization of this isolate revealed a plasmid carrying both cephalosporin and fluoroquinolone resistance genes which is uncommon in Salmonella in the United States. To our knowledge, this is the first time this combination of resistance genes has been found in NARMS retail meat testing and raises concern since ceftriaxone (a cephalosporin) and ciprofloxacin (a fluoroquinolone) are critical drugs to treat severe Salmonella infections (CDC, Antibiotic Resistance Threats in the United States, 2019).
MDR profile of the NARMS isolate 19MN11PC02
| Class | Antimicrobial | Resistant | Genes |
|---|---|---|---|
| Aminoglycosides | Gentamicin | X | aac(3)-IIa |
| Aminoglycosides | Streptomycin | X | aadA1, aph(3')-Ia, aph(3'')-Ib, aph(6)-Id |
| Beta-lactam combination agents | Amoxicillin-Clavulanic Acid | ||
| Cephems | Cefoxitin | ||
| Cephems | Ceftriaxone | X | blaCTX-M-15 |
| Folate pathway antagonists | Sulfisoxazole | X | sul2 |
| Folate pathway antagonists | Trimethoprim-Sulfamethoxazole | X | dfrA14 |
| Macrolides | Azithromycin | ||
| Penems | Meropenem | ||
| Penicillins | Ampicillin | X | blaCTX-M-15, blaOXA-1, blaTEM-1 |
| Phenicols | Chloramphenicol | X | catA1, catB3 |
| Quinolones | Ciprofloxacin | X | aac(6')-Ib-cr5, qnrB1 |
| Quinolones | Nalidixic Acid | X | qnrB1 |
| Tetracyclines | Tetracycline | X | tet(A), tet(B) |
Fast Facts
Isolate: 19MN11PC02.
Organism: Salmonella I 4,[5],12:i:-
Source: Retail pork chop
Location(s): MN
Date collected: November 2019
Additional information on genetically similar Salmonella I 4,[5],12:i:- from pork and human clinical specimens can be found at NCBI’s Pathogen Detection Portal. At this time, the resistance pattern in Salmonella I 4,[5],12:i:-reported here is considered a rare finding in the United States.
Over the past decade, Salmonella I 4,[5],12:i:- has emerged as a major public health threat in Europe and the United States. In 2019, it ranked fifth among laboratory diagnosed nontyphoidal Salmonella infections in the United States (CDC).
Next Steps
FDA is performing additional genomic testing to better understand the origins of genes that caused the extensive resistance profile in 19MN11PC02 and other related I 4,[5],12:i:- isolates.
Consumer Safety
This notification is to inform stakeholders of this novel finding. While this rare finding by itself does not signify elevated risk, the public is advised to continue practicing safe handling of raw meats, as recommended by the USDA, and proper hygiene around animals as recommended by the CDC.

![Number of MDR Salmonella I 4,[5],12:i:- isolates with azithromycin resistance gene mph(A) tested by NARMS, by isolation year, January 1, 2017–May 29, 2024 The number of MDR Salmonella I 4,[5],12:i:- isolates with azithromycin resistance gene mph(A) tested by NARMS 1/1/2017-5/29/2024. In Humans, in 2017, there were 3 isolates; in 2018, there were 6; in 2019, there were 3; in 2020, there were 4; in 2021, there were 13; in 2022, there were 20; in 2023, there were 47; in 2024, there were 16. In food, there was 1 in 2020, 2 in 2023, and 3 in 2024. In animals, there was 1 isolate in 2019, 1 in 2020 and 2021, and 2 in 2022 and 4 in 2023.](/files/Graph-1-isolates-with-azithromycin.jpg)
![Percentage of Salmonella I 4,[5],12:i:- isolates with azithromycin resistance gene mph(A) that were predicted resistant, by antimicrobial (n = 127), as of May 29, 2024 — NARMS This graph show the percentage of Salmonella I 4,[5],12:i:- isolates with azithromycin resistance gene mph(A) that were predicted resistance, by antimicrobial, as of May 29,2024. The first bar in grey, show the percentage of XDR isolates with the mph(A) genes. The remaining bars in blue, show isolates resistance to other antimicrobials recommended for empiric and alternative antimicrobials.](/files/Graph-2-percent-of-Sal.jpg)